Example 10: Analyzing the Results of the High-Lat Run¶
In this Example we analyze the results of an MPI run of NPTFit performed over high latitudes.
The example batch script we provide, Example10_HighLat_Batch.batch
is for SLURM. This calls the run file Example10_HighLat_Run.py using
MPI, and is an example of how to perform a more realistic analysis using
NPTFit.
NB: The batch file must be run before this notebook.
NB: This example makes use of the Fermi Data, which needs to already be installed. See Example 1 for details.
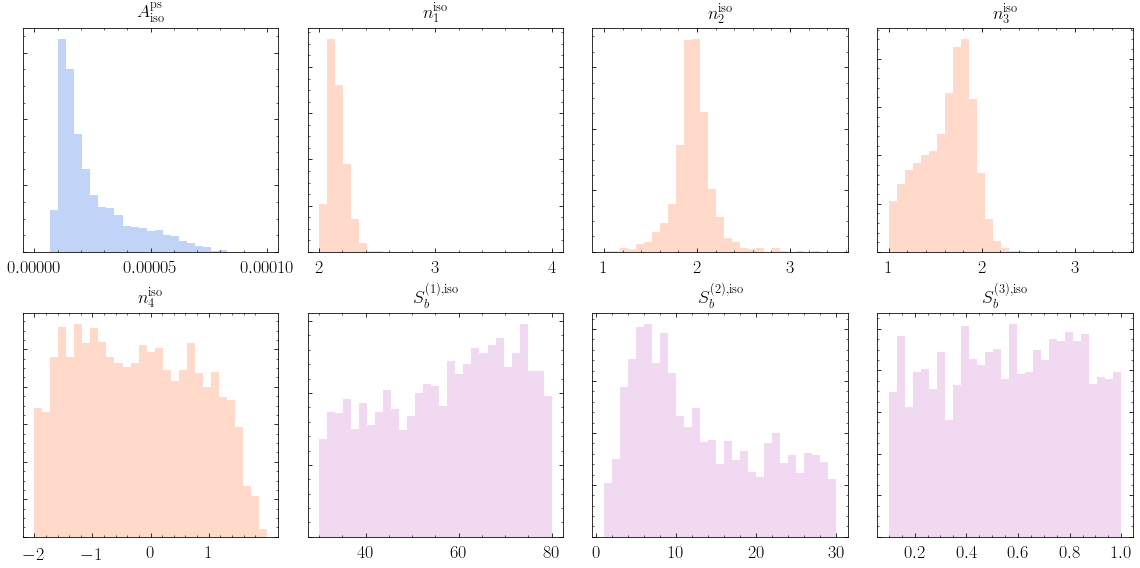
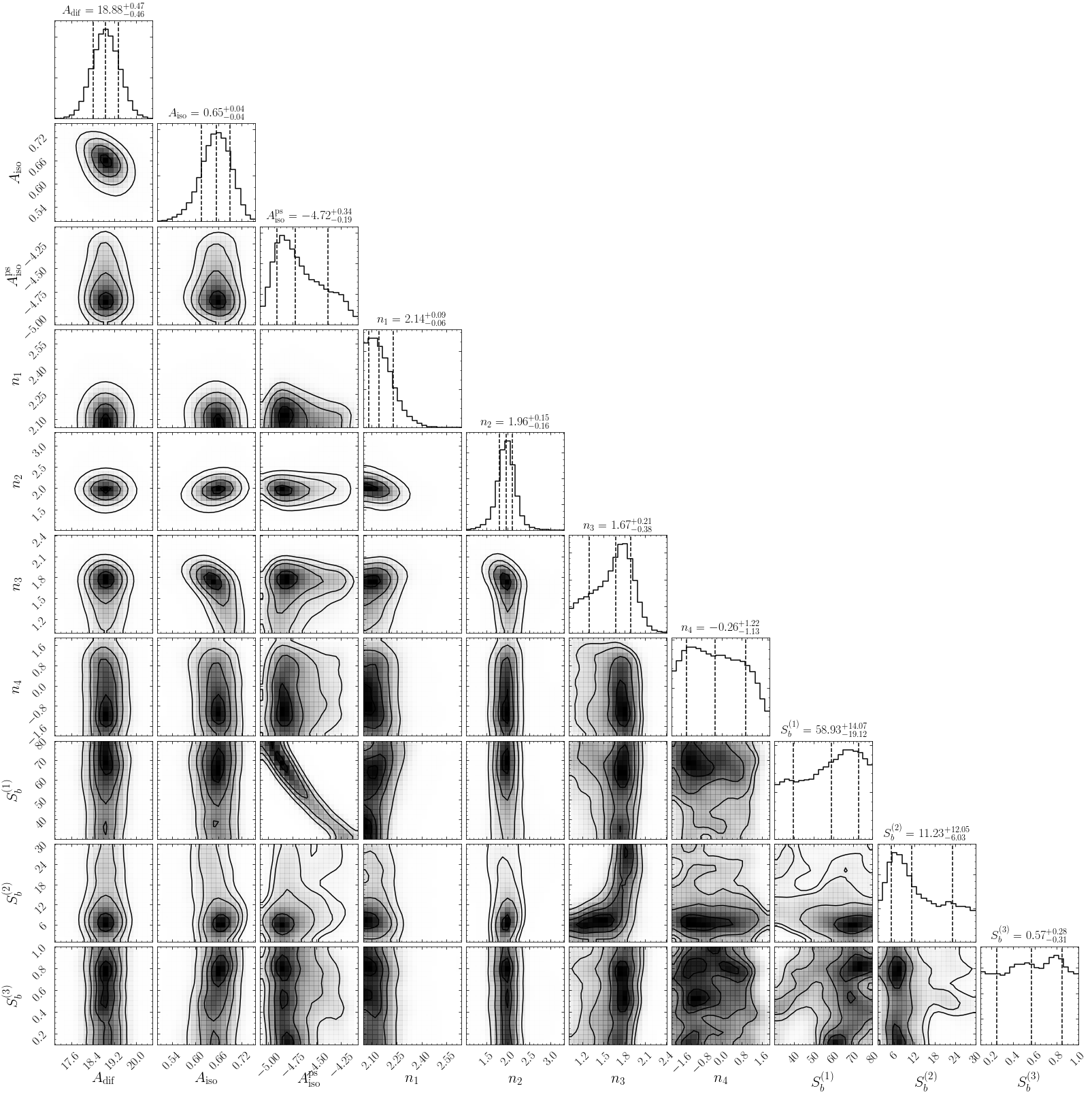
In this example, we model the source-count function as a triply-broken power law. In detail, the source count function is then:
and is thereby described by the following eight parameters:
This provides an example of a more complicated source count function, and also explains why the run needs MPI.
Load in scan¶
We need to create an instance of nptfit.NPTF and load in the scan
performed using MPI.
Loading the psf correction from: /group/hepheno/smsharma/NPTFit/examples/psf_dir/gauss_128_0.181_10_50000_1000_0.01.npy
The number of parameters to be fit is 10
Finally, load the completed scan performed using MPI.
analysing data from /group/hepheno/smsharma/NPTFit/examples/chains/HighLat_Example/.txt
Analysis¶
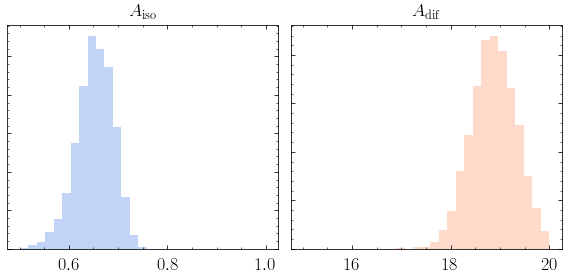
As in Example 9 we first initialize the analysis module. We will provide the same basic plots as in that notebook, where more details on each option is provided.
1. Make triangle plots¶
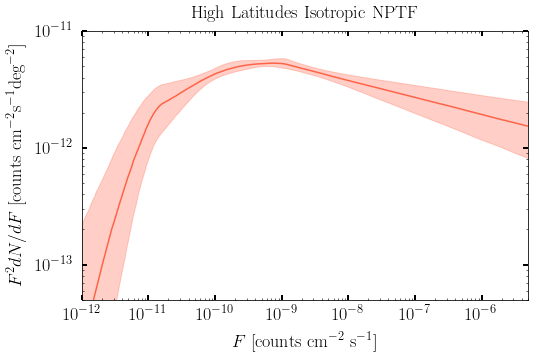
2. Get Intensities¶
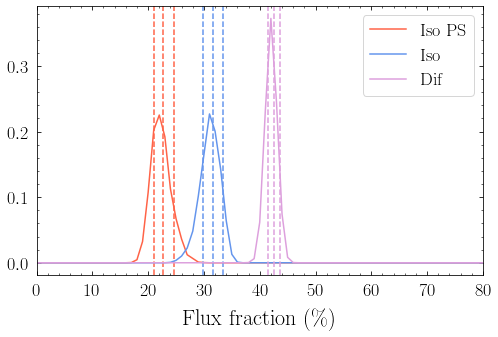
Iso NPT Intensity [ 1.21483730e-07 1.31199062e-07 1.41996475e-07] ph/cm^2/s
Iso PT Intensity [ 1.40131525e-07 1.48859129e-07 1.56973582e-07] ph/cm^2/s
Dif PT Intensity [ 1.96440673e-07 2.01367819e-07 2.06396272e-07] ph/cm^2/s
4. Plot Intensity Fractions¶